The Next Salvo In The Diagnosis Wars
OpenAI's "agentic" diagnostic tool.
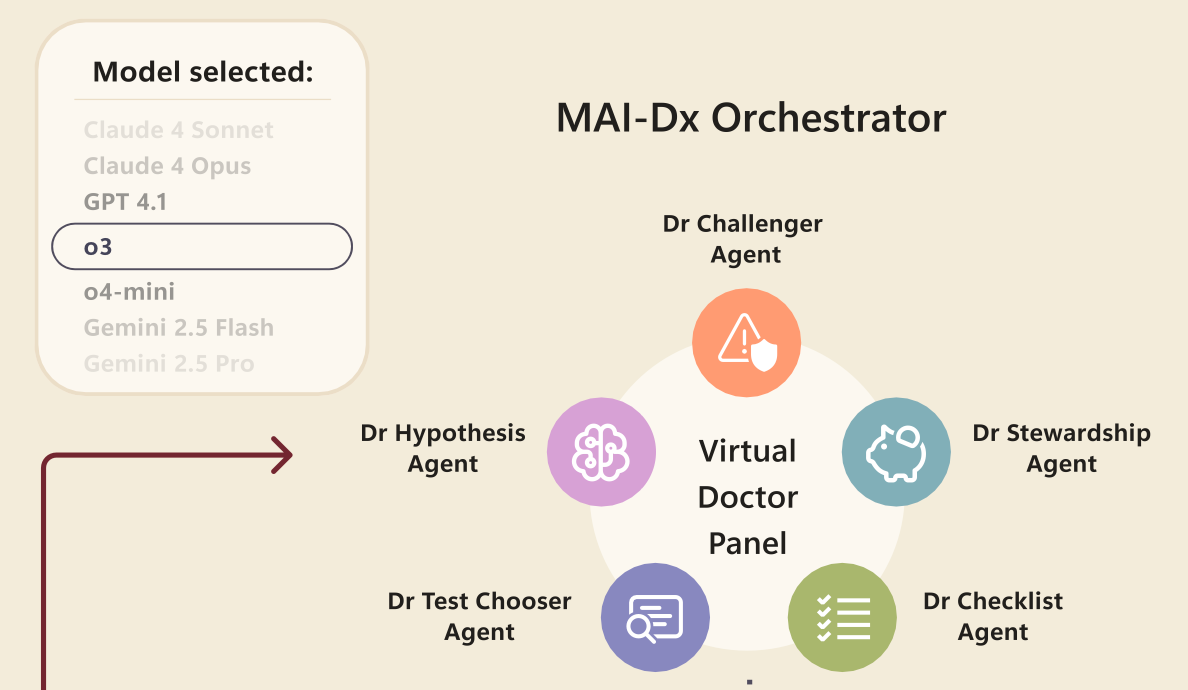
This is OpenAI + Microsoft Health’s contribution to the diagnostic wars in medicine, the “MAI Diagnostic Orchestrator”. The MAI-DxO is an agentic structure superimposed upon general large language models, the purpose being to coordinate its actions among five worker bees:
The agentic structure is optimised for GPT-4.1, but, because each agent does little more than prompt an underlying general purpose LLM, the MAI-DxO is platform agnostic.
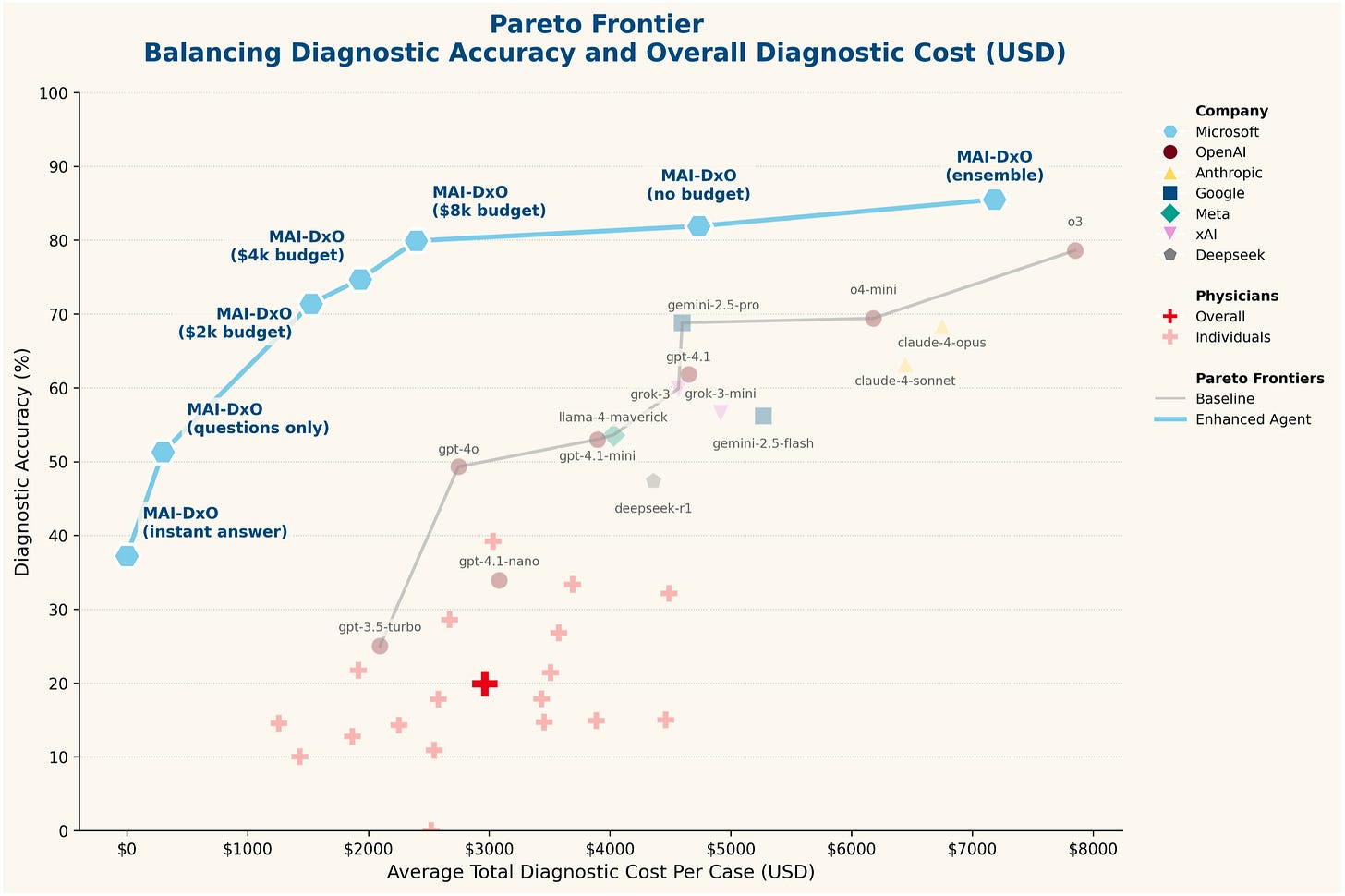
The oft-repeated performance demonstration is here, as performed on their chain-of-thought-style benchmark created from New England of Medicine Journal Case Challenges:
Part of their goal, as you can see, is also to determine the cost-effectiveness of the diagnostic strategy – attempting to replicate the “real world” constraints facing clinicians.
Naturally, the agent wins! This figure is a bit misleading, however, as this comparison shows the agent against interactions with general purpose models using a baseline prompt. Then, scattered below, is “human performance” – abysmal! Well, it’s abysmal because the NEJM case challenges are frequently esoteric cases requiring multi-specialty input, and the humans here are hamstrung by not even being allowed access to any reference texts or online resources. Much will be made about the relative superiority of LLMs over humans, but this is an inadequate strawman comparison.
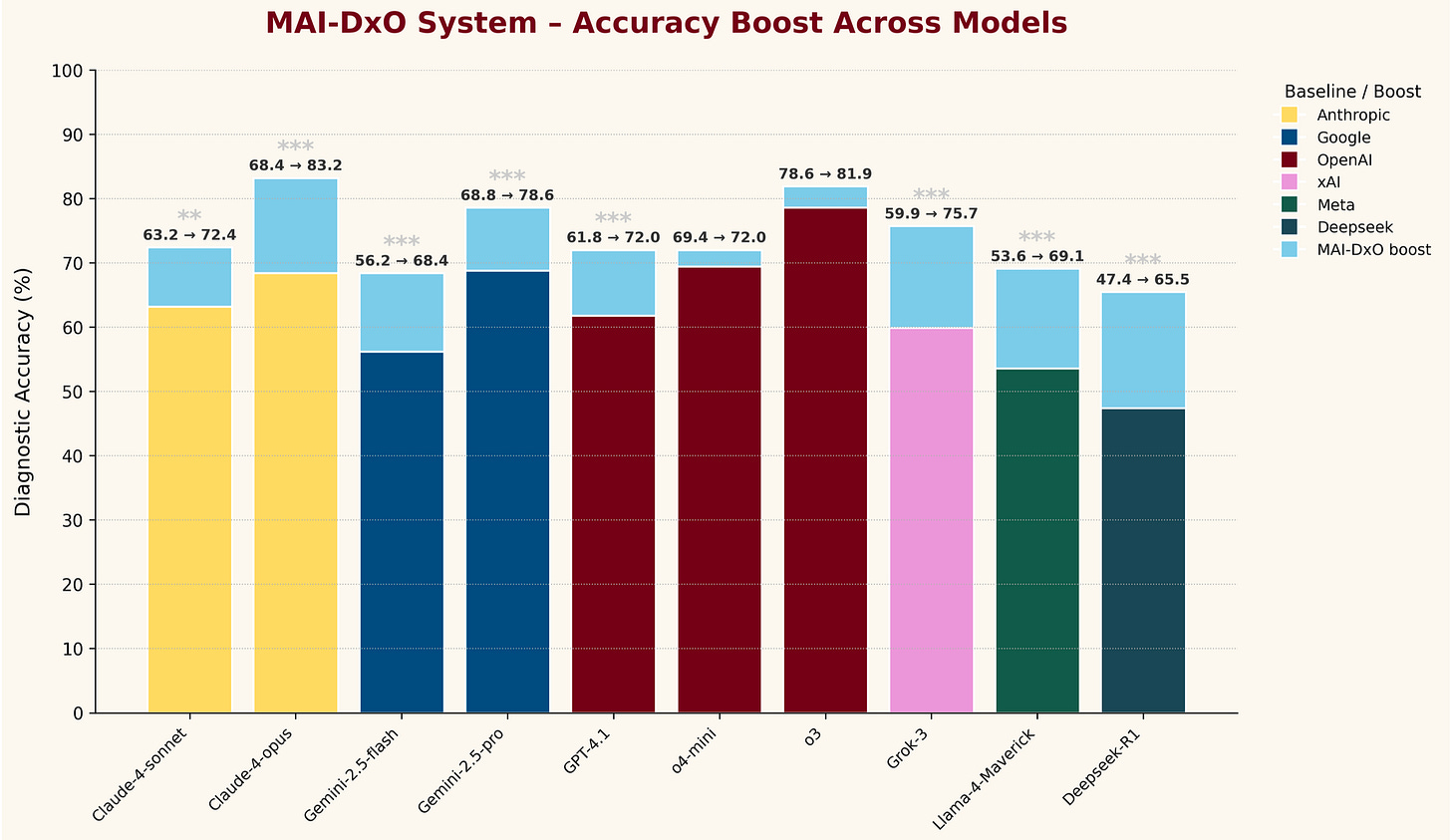
A little bit more interesting is their “what does MAI-DxO add to other models” figure:
The differences are mostly trivial, but it is interesting to see the top Claude, Gemini, and o3 models all hit similar performance levels when integrated into this agent.
Obviously, there’s a performance/energy cost to marrying multiple frontier LLMs – potentially outweighed by effective clinical application, as long as the performance advantage to the agentic structure is substantial. Then, it would be interesting to compare this to one of the medically-optimized models, or to overlay this agent on top of them. Finally, because they’ve invented their own specific diagnostic testing structure, there’s not really any way to compare these results to any other benchmarking.
These are also complex cases with interesting pathology, and not representative of the vast majority of clinical medicine. We certainly can’t be ordering “Ultrasound-guided core biopsy of the right peritonsillar mass (large-bore), send for routine H&E, immunohistochemistry (CD31, D2-40, CD34, ERG, GLUT-1, pan-cytokeratin, CD45, CD20, CD3, Ki-67)” on every sore throat – nor does this generalize to health systems without access to such advanced pathology testing.
Another interesting step on the journey, regardless.

Maybe I'm missing something, but if these diagnostic models are just calling general LLMs, isn't NEJM exactly the kind of input the LLMs would be trained on, potentially including the diagnostic cases they are testing against? I know I've seen some of these papers where they intentionally created test scenarios that couldn't have been included in the training material but that doesn't seem to be the case here.
Even if they are protecting from that particular source of confounding, it seems like the more esoteric/rare the disease the more likely both your training and testing materials are to consist of a small number of case reports and derivatives of those case reports. As a result, I'd expect the model to perform well against those testing materials but see no reason to think it would perform nearly as well on an actual patient presentation.